Livermore’s Forensic Science Center is helping law enforcement agencies by developing the first-ever protein-based biological identification method.
Improving Law Enforcement Tools
Law enforcement and other federal and state agencies working in forensics depend on scientific and statistically valid methods to identify individuals. To date, DNA fingerprinting has been the only forensic method that demonstrates a high degree of statistical certainty in connecting evidence to a specific individual or source. By developing the first-ever biological identification method that uses protein found in hair, skin, bone, and teeth, a team at Livermore’s Forensic Science Center (FSC) is strengthening and expanding forensic tools available to law enforcement. This protein marker identification technology could be used when DNA samples are compromised or not available.
A Complement to DNA Methods
High-quality DNA profiling can narrow down the identity of an individual to one person worldwide. However, DNA is fragile by nature, thus this technique is limited in scope. Successful profiling depends on the presence of a DNA template (of sufficient quantity and quality) that can be amplified using polymerase chain reaction to produce information about the sample’s genotype. Unfortunately, exposure to biological environments and chemical processes can reduce DNA’s template length and modify its base structure.
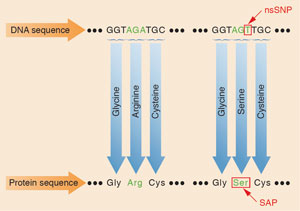
In contrast, proteins are more stable, abundant, and environmentally persistent than DNA, making protein marker identification technology a complementary or alternative solution. The markers that can differentiate one person from the next are protein sequences containing single amino acid polymorphisms (SAPs). These altered amino acids are genetic variations generated from nonsynonymous single nucleotide polymorphisms (nsSNPs)—DNA mutations that change a protein’s amino acid sequence.
Quantifiable Results
Using protein marker identification technology, FSC scientists can analyze just a single strand of hair (o r a small sample of skin, bone, or tooth) and determine the probability of how unique that profile is in the general population. First, the sample is broken down into peptides and analyzed using liquid chromatography mass spectrometry. Scientists then use software to screen the data for genetically variant peptides (GVPs), small segments of proteins that contain SAPs. The results are compared to FSC’s SAP database which includes nsSNP protein markers that are candidates to be included in a “human identity” panel. The panel—which will include 100 of the nsSNP protein markers from the database when complete—will serve as a “checklist” of the protein markers that need to be screened in each sample to narrow down the identity of the sample’s donor. The scientists are looking for matches between the sample and the protein marker in the panel. With data on the frequencies of each matching nsSNP in the population, the team of scientists can estimate the probability that the same marker profile exists in a given number of people.
As the number of matches found between nsSNP markers in the sample and the human identity panel increases, so does the power of discrimination—that is, the more certain the team is regarding the identity of the sample’s donor. A panel of 100 protein markers, for example, could narrow down the identity of the donor to one in 10 billion—essentially, one person worldwide. Within two years, FSC expects to develop a kit containing sample preparation material and synthetic examples of protein markers in the panel for law enforcement and other Laboratory agencies to use so they can determine what to look for when examining a sample.
Looking Ahead
In addition to building a database of nsSNP markers that are fairly common in humans, FSC scientists are exploring rare mutations that will provide authorities with a greater ability to differentiate individuals. This capability could be useful to criminal investigators who are trying to distinguish individuals from complex biological mixtures, which can be challenging with DNA profiling. For example, law enforcement might want to know whether a specific person was in a particular room at a certain time and whether he or she touched a particular object. Forensic scientists could look for protein markers in the skin or hair the person left behind. FSC is also developing a technique in which both mitochondrial DNA and proteins can be extracted from a single hair.
This new technology represents how FSC is transforming forensics from traditional subjective methods (which rely on an expert’s opinion when detecting matches by examining hair or fingerprints under a microscope) to advanced objective methods. In addition to broadening what types of samples can be used for analysis, FSC methods can also quantify just how often a particular sample appears in the population.
Initial funding for this project was provided by the Department of Defense. Ongoing support continues through Livermore’s Laboratory Directed Research and Development Program and the Intelligence Advanced Research Projects Activity.