Quick, Accurate Pathogen Detection Enhances Public Health and Biosecurity
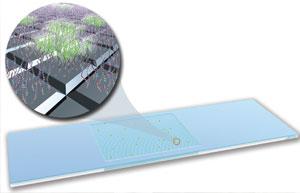
The Lawrence Livermore Microbial Detection Array (LLMDA) can identify thousands of different microbes within 24 hours for faster diagnosis and treatment of biological threats.
Time Is of the Essence
Diagnostic platforms that can rapidly detect and characterize microbial agents—the viruses, bacteria, and fungi that cause disease— are becoming increasingly important to safeguard human health and safety. Infectious diseases pose a growing threat to public health because of population growth, international air travel, bacterial antibiotic resistance, and other factors. In addition, forensic experts are increasingly concerned that pathogens, perhaps genetically engineered, could be released deliberately by terrorist organizations or rogue states.
Although several techniques exist for identifying pathogens via their genetic code, such as polymerase chain reaction (PCR), most of these methods are too costly or slow to efficiently analyze clinical and environmental samples that may contain hundreds or even thousands of different microbes. The Lawrence Livermore Microbial Detection Array’s (LLMDA’s) DNA-based detection provides 50- to 100-fold greater coverage of microbes compared to traditional culturing techniques and does so at a much faster rate, hours rather than days. LLMDA provides the first high-throughput, cost-effective platform for detecting a broad range of possible pathogens in a short amount of time, potentially saving lives.
Tech Serves Multiple Users
LLMDA uses probes consisting of short stretches of RNA or DNA to help uniquely identify pathogens contained in a sample. These probes are selected using a set of algorithms developed by Livermore bioinformaticists, including an automated composite likelihood maximization algorithm to process output data. The system was designed to detect any known microorganism sequence, based on all available genome sequence data. Hundreds of thousands of probes are designed for more than 12,000 microbial species from 4,776 viruses, 6,906 bacteria, 370 archaea, 143 protozoa, and 414 fungi. LLMDA also has the potential to detect novel, divergent species by incorporating probes that are conserved at the family taxonomic level.
In trials for government agencies, international researchers, and health-product companies, LLMDA has accurately and rapidly identified bacterial and viral pathogens present in human and animal clinical, environmental, and product samples. LLMDA is also being applied to identify viruses and bacteria that are correlated with high cancer risk, vaccine safety, and biosecurity. If widely adopted, LLMDA could allow professionals in medicine, pharmaceuticals, law enforcement, product and food safety, public health, animal health, the military, and global disease surveillance to detect within 24 hours any virus or bacteria that has been sequenced, allowing for faster diagnosis and treatment of disease.
Helping Heal Soldiers’ Wounds
In times of war, a military medical team responding to an infantry soldier with extensive wounds must quickly deduce the most effective way to treat the victim. Typically, clinicians determine a wound-care approach based upon general intuition and experience that includes considerations such as how and what bacteria in the wound could help or hinder the patient’s overall recovery.
In collaboration with investigators from the Naval Medical Research Center, Walter Reed Army Institute of Research, Uniformed Services University of the Health Sciences, and University of California at Davis, Livermore researchers used LLMDA to analyze more than 100 wound samples from 44 U.S. soldiers injured in Iraq and Afghanistan. The system was sensitive enough to detect bacteria such as Pseudomonas and Acinetobacter baumannii in many of the wounds that failed to heal correctly.
The results of the study ultimately indicated that different pathogens can positively or negatively affect wound outcomes. Researchers also determined that certain pathogens important to wound healing may not be identifiable by standard culture- based detection methods. LLMDA’s success in identifying bacterial pathogens and their role in aiding or impeding wound healing can assist medical professionals in personalizing treatment and accelerating recovery for patients, whether soldiers or civilians.
Insight into Carcinogenic Pathogens
Infection-associated cancers are on the rise worldwide due to shifting pathogen habitats, a more interconnected world, and an aging population with a longer life expectancy. LLMDA has been applied in several studies designed to help identify whether particular pathogens could be associated with the development of specific cancers.
In one study, in which LLMDA sequenced specimens from lung cancer patients, results showed the consistent presence of viral DNA from retroviruses, human papillomavirus, and several other viruses in a large percentage of the specimens. In another study, LLMDA accurately detected a Kaposi’s sarcoma-associated herpesvirus in specimens derived from patients with bladder cancer, indicating a possible correlation between the pathogen and cancer development. A third study implemented LLMDA to screen human lymphoma tissues, confirming the presence of Epstein–Barr virus with no false positive results. Thus, LLMDA is proving to be a sensitive and alternative diagnostic method for identifying known viral pathogens associated with tumors.
Ensuring Vaccine Safety
Vaccines made from highly effective, safe, and relatively inexpensive, live-attenuated viruses, protect against numerous human and animal viral infections. However, attenuated live vaccines, such as those for chicken pox and measles, also carry a potential risk of contamination with adventitious viruses introduced during the attenuation process, from the cell lines used, or from animal sera or other biologics contained in cell cultures.
LLMDA has been applied to screen samples extracted directly from vaccines and from patients who were recently vaccinated or were suffering from other conditions that could be associated with receiving a contaminated vaccine. The tests were done to confirm the presence of bacterial and viral pathogens as well as adventitious agents. In both studies, LLMDA proved that viral metagenomics and microarray technology could be used as an effective surveillance tool for interrogating current and future vaccines for genetic variants of the attenuated viruses and the presence of adventitious viruses to enhance confidence in the safety of the vaccine supply.
Better Detection of Animal Pathogens
New infectious diseases in animal food production systems can have enormous impacts that affect domestic consumption and exports. Such diseases can also negatively affect public health, especially in cases where the pathogens move from animals to humans. In collaboration with Kansas State University, Livermore researchers applied LLMDA to evaluate the impact and association of microbiomes— all microorganisms in a particular environment—connected with two of the most devastating viral diseases in swine. During their yearlong study, researchers evaluated known and unknown microbes in blood, oral fluid, and tonsils from pigs that have co-infections of porcine reproductive and respiratory syndrome virus and porcine circovirus-2. LLMDA easily identified both pathogens, but at higher concentrations compared to gold-standard PCR detection methods. In addition, co-infection could be identified with a single sample. In the future, LLMDA could be adapted for cattle and poultry diagnostic tests, as well as for companion animals, such as dogs and cats. On a larger scale, it may also help the U.S. detect the emergence of foreign animal diseases at their outset to prevent major disease outbreaks.
Ancient Remains Reveal Disease
Scientists studying the origins of infectious disease and population health over time normally find that ancient human remains contain highly degraded DNA in which the pathogen represents only a tiny fraction of the total DNA yield. Lengths of these fragments are often as small as 30 to 50 bases compared with regular DNA fragments that are hundreds to thousands of bases long, which makes them difficult to detect with PCR or sequencing techniques. LLMDA technology is both five- to ten-times faster and less expensive than current genomic sequencing for studying pathogens in ancient DNA.
As part of a study conducted by researchers at McMaster University and Lawrence Livermore, LLMDA identified human bacterial pathogens from two separate archaeological remains. An intestinal sample showed the presence of cholera preserved from a patient who died from the disease during an 1849 outbreak in Philadelphia. The other sample indicated the presence of Yersinia pestis (the causative agent of plague) in a medieval tooth dating back to 1348. The amounts of pathogen in the two samples tested were minute—three ten-thousandths of the total sample for cholera and eight ten-thousandths for Y. pestis.
LLMDA selectively targets pathogens of interest and works well even with heavily degraded DNA, typical of most fossil and archival remains. Research studying ancient human remains for infectious disease can offer clues as to how past pandemics happened and why they were so deadly. Thus, characterization of ancient pathogens will help scientists better understand pathogen evolution and help predict future emerging threats.
System Bound for Space
In a joint effort with researchers from three National Aeronautics and Space Administration (NASA’s) agencies—Jet Propulsion Laboratory, Ames Research Center, and Johnson Space Center—Livermore scientists are using LLMDA to study the microbiome associated with the International Space Station and its crew. By characterizing microbes in the air, on the surface of the station, and among the astronauts, scientists gain a better understanding of whether any pathogens exist that might be harmful to the crew’s health.
As part of this project, LLMDA will detect microbes in the DNA samples—mouth, saliva, and skin specimens—obtained from the crew members. After the study is completed, NASA may consider miniaturizing LLMDA or a similar instrument for use on deep space missions requiring human habitation, such as a potential long-term exploration mission to Mars. The crew could use the system before consuming food, drinking water, or working in a closed area as a preventative measure. Also, if a crew member becomes ill, LLMDA could help determine whether the cause of the sickness is bacteria, fungi, or viruses, enabling NASA flight surgeons to administer the treatment based on the pathogen.
Higher Throughput at Lower Cost
LLMDA has proven to be a highly effective research tool in the formats supplied by Roche NimbleGen and Agilent, which incorporate DNA probes onto glass slides. However, the cost per sample of these formats has been too high to foster regular production usage. To overcome this challenge, Livermore has been working under a Cooperative Research and Development Agreement (CRADA) with Affymetrix, a leading microarray company now part of Thermo Fisher Scientific, Inc., to develop a higher throughput version of LLMDA that achieves a lower cost per sample.
The Applied Biosystems Axiom Microbiome Array is now commercially available in both 96- and 24-sample formats. Each of the 96 arrays contains 1.4 million probes targeting the same 12,000 microbial species as the Agilent version. The increase in probes significantly reduces the background “noise” of false-positives. The cost per sample for in-house processing with the Thermo Fisher Affymetrix GeneTitan instrument is comparable with multiplex PCR assays. As a result, the higher throughput version of LLMDA can compete with multiplex PCR and DNA sequencing for many important applications in public and animal health, food and product safety, biosurveillance, and biodefense.
Crucial Tech for Better Care
Because early detection and unmistakable pathogen identification are crucial to limiting the potentially catastrophic human and economic costs of a bio-attack, LLMDA provides a method to rapidly and accurately identify pathogens that could be used to sicken or kill urban populations, livestock, or crops. As a diagnostic tool, the device could also realize significant cost savings in routine health care.
The device is fully updatable, allowing for the incorporation of newly sequenced organisms and strains in subsequent versions of the array. In addition, LLMDA’s Web-based data entry is an essential characteristic for a deployable pathogen-detection technology. With its unique capabilities, LLMDA is poised to provide a powerful new weapon to fight disease, ensure the safety of vaccines and food products, and provide increased protection from a bio-attack.